| Biography | Research | Publications | Teaching | Group | Projects | Software | Sponsors | Collaborators |
| PROJECTS |
|
Model
Pipeline
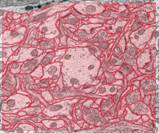
Electron
microscopy is used to reveal the structure of brain tissue at the micron and
submicron scales. However, the ability to analyze the
spatial relationships between various cellular structures as well as the
arrangement of organelles within cells is limited in two-dimensional electron
microscopy images. Reconstruction from serial section transmission electron
microscopy (ssTEM) offers the possibility of recreating neuronal and glial
processes and their organelles in spatially realistic three-dimensional models.
Our EM data comes from area CA1 of the hippocampus (courtesy of Dr. Kristen
Harris). We
outline the overall computational framework that we have been developing for
converting imaging data to spatially realistic meshed models above. There are
four major stages: 2-D image processing (white box), 2-D geometry processing
(blue box), 2-D to 3-D reconstruction (yellow box), and 3-D geometry processing
(green box). An additional set of procedures, the reducing procedures (orange
box), are used to generate 1-D multi-compartment models suitable for NEURON
from 3-D reconstructions. The red boxes in Fig. 2 represent components of
ongoing development.
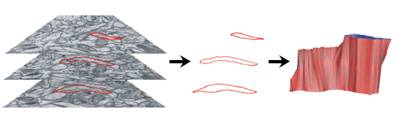
The image above shows several stages of our
processing pipeline. The boundaries of all cellular elements (dendrites, axons,
and glia) are traced and classified in each 2-D image slice (Fig. a). These
contours undergo further processing steps (referred to as 2-D curation
procedures) that accomplish such goals as removing boundary overlaps and
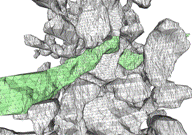
resampling by spline-fitting. The 2-D contours are tiled into 3-D objects (Fig.
d) using our ContourTiler algorithm (implemented in our VolRover software
package). This provides us with 3-D spatially realistic reconstructions of
dendrites (Fig. e), axons, and glial processes within a reconstructed neuropil.
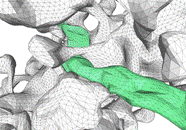
However, the quality of the initial, tiled surface mesh (Fig. f) is not
sufficient to be useful for computational simulations. Our set of 3-D curation
procedures removes boundary overlaps in 3-D and applies mesh quality
improvement algorithms, such as smoothing, decimation (reducing the number of
triangles), and fitting by algebraic spline functions, in order to generate a
forest of non-intersecting, high quality-meshed neuronal (Fig. 3g) and glial
processes. These reconstructions are then ready for use in simulations of
neuronal activity. | |||||||||